Introduction
Oral biofilms contains over 700 bacterial species on the soft and hard tissue in the mouth. Oral biofilms forms dental caries, which lead to an inflammatory host response around the gingival crevice. In Streptococcus mutans, a biofilms is formed on the surface of the tooth by host-pathogen interaction, and the early colonists attach to the tooth enamel and begin to form a three-dimensional biofilms. In the early stage, oral biofilms includes members of Streptococcus, Actinomyces, and Haemophius. Streptococcus mutans resides primarily in biofilms that form on the tooth surfaces as a major etiological agent of dental caries. Streptococci can promote the establishment of subgingival biofilmss and change the structure of subgingival biofilms in the absence of an early colonizer [1,2]. The subgingival has obligate anaerobic bacteria such as Actinomyces and Prevotella species higher than Gram-positive bacteria. Prevotella intermedia has been implicated in the development of chronic oral infection. Co-cultures of F. nucleatum and P. intermedia strains increased biofilms formation compared to single cultures. Physical contact between the F. nucleatum strain and P. intermedia aggregation plays an important role in the formation of biofilms by the strain [2,3]. F. nucleatum is one of the first Gram-negative species to become established in bioflm and central species in physical interactions between Gram-positive and Gram-negative species that are likely to be important in biofilms colonization [4].
To reduce oral bacteria number, oral biofilms can be eliminated by an artificial or chemical way. As a chemical removal method, there is the administration of enzyme, antibiotic, or antimicrobial. The representative chemical medicine as a mouth rinses type is Chlorhexidine (CHX), which has outstanding efficacy on supragingival plaque controls [5]. CHX is effective to restrain both Grampositive and Gram-negative bacteria, and reduce the attachment of Porphyromonas gingivalis on epithelial cells [5,6]. Benzethonium chloride (BC) is able to reduce dental plaque formation [7]. If Listerine is used for 60 sec twice a day, it has considerable efficacy on lessening plaque and restraining gingivitis and gingival bleeding [8]. Laine et al. [9] reported that if Amine fluoride-stannous fluoride mouth rinses have been used, gingival bleeding index could be decreased. If gargling water (mouth rinse), BC, CHX, or essential oil (EO) are treated on planktonic cells, it has over 99% antibacterial effects on periodontitis and cariogenic Streptococcus mutans. Besides, 0.1% Chlorhexidine is indicated as restraining the formation of oral biofilms up to 68% [5,10].
So far, studies have been conducted about the efficacy of mouth rinses on a planktonic cell or single bacterium; therefore, in this study, assessed the antibacterial effect of mouth rinse solution against multispecies oral biofilms consisting of S. mutans, F. nucleatum and P. intermedia. We purchased products on the domestic market and confirmed antibacterial effects of each mouth rinse component on multiple oral biofilms in vitro. We measured APT and CFU to confirm the presence of viable cells, morphology visualized by SEM and gene expression conformed RNA sequencing.
Methods
1. Bacterial strain
Streptococcus mutans GS-5, Fusobacterium nucleatum ATCC 15037 and Prevotella intermedia ATCC 49046 were used in this study. F. nucleatum ATCC 15037 and P. intermedia ATCC 49046 were grown on Tryptic soy agar (TSA, Difco Laboratories, Detroit, Mich) plate contain 5% sheep blood, 5 μg/ml hemin and 1 μg/ml Vit K1. S. mutans were grown on Brain Heart Infusion (BHI, Difco) plate [11]. All bacterial strains were grown BHI broth and incubated at 37℃ anaerobic conditions (85% N2 , 10% H2 , 5% CO2 ).
2. Mouth rinsing solution treatment of preformed multiple oral biofilms
In this study, mouth rinsing solutions were selected that can be easily purchased in Korea then pH was determined on each solution in duplicate by pH meter (Mettler toledo, Switzerland). Saline was used as control. The manufacturers, ingredients, and pH of the mouth rinse solutions are shown in (Table 1). The bacterial culture was grown to exponential phase, and then diluted in fresh medium to an optical density at 600 nm (OD600) of approximately 0.1. To prepare the multispecies suspension. Optical density (OD)-adjusted S. mutans, F. nucleatum and P. intermedia were mixed in equal proportions. The culture was performed by adding 200 μl of cell suspension to one well of a polystyrene 96-well. After 48 h of incubation at 37℃ anaerobic conditions, planktonic bacterial cells were removed by aspiration. The cell washed twice with saline. 100 μl of each mouth rinsing solution was added to each well, the plates were incubated for 1 min at room temperature according to the manufacturer’s instructions, rinsed thoroughly and repeatedly with saline.
3. ATP analysis
ATP bioluminescence was measured using the BacTiter-GloTM microbial cell viability assay (Promega Corp., Madison, WI, USA) according to the manufacturer’s instructions. After preformed biofilms treated mouth rinsing solution, the cell washed twice with saline. After the cell suspension with 100 μl saline, cell transferred into a new white 96 well microplate. Afterwards, add equal to the volume of BacTiter-GloTM reagent in each well. The sample incubated for 5 min form room temperature for the traction time. All ATP measurements were done in duplicates, the relative light units (RLU) were measured in a luminometer.
4. Colony forming unit (CFU)
After exposure to the mouth rinsing solution, the cell washed and suspended with saline. The samples were subjected to 10 fold serial dilutions in BHI broth and plated onto TSA agar and incubated at 37℃ for 7 days under anaerobic conditions as described above. The colonies were counted.
5. Mouth rinsing solution treatment of preformed biofilms on saliva-coated hydroxyapatite (HA) disk
Hydroxyapatite (HA) disks (φ=13 mm×3 mm) were soaked in 3 mL of the clarified whole human saliva contain 0.04% NaN3 at room temperature overnight as described previously [12,13]. Mixed all bacterial species was transferred saliva coated HA (sHA) disk placed into the well then incubated at 37℃ for a week anaerobic conditions, with BHI broth changed everyday. After a week, sHA disk were transferred to a new plate and washed twice with saline to remove unattached bacteria. Preformed oral biofilms on sHA disk treated EO (Johnson & Johnson, Thailand) and saline to control for 5 min at room temperature.
6. Scanning electron microscope (SEM)
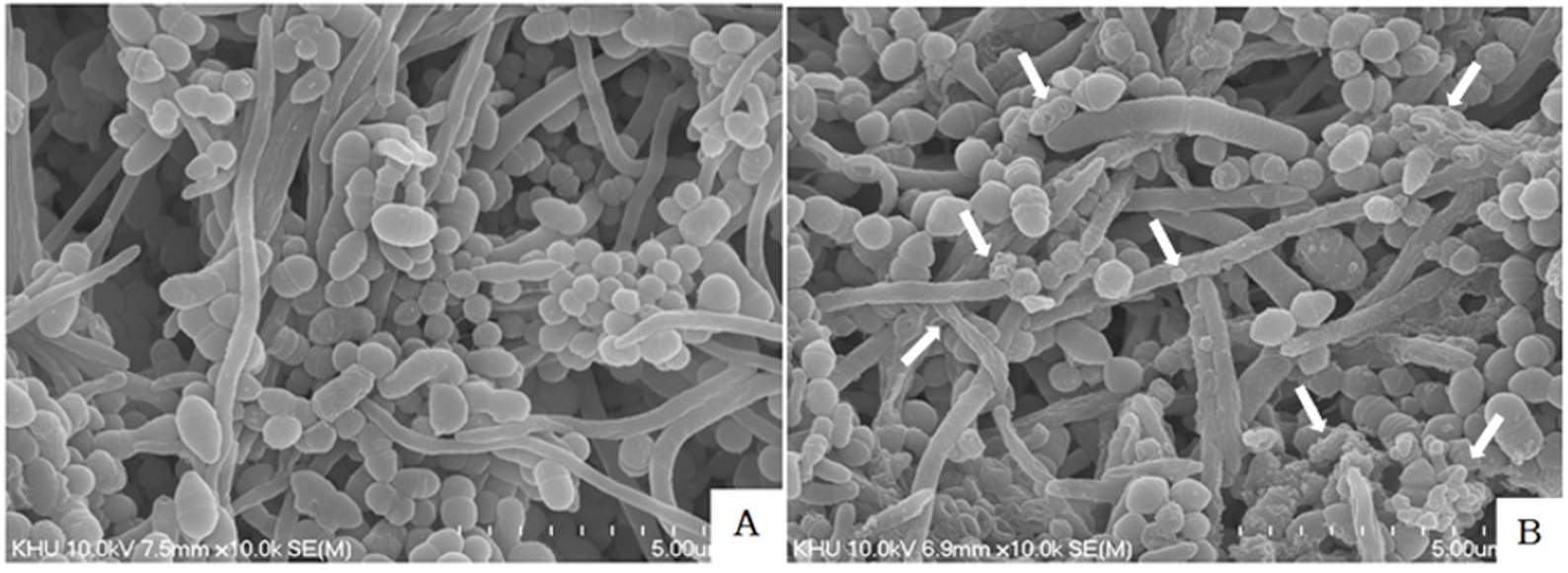
As being visualized after treated to the solutions for 5 min on the preformed multiple oral biofilms sHA disks were observed by SEM at a magnification of 20,000×operating at 5 kV. The preformed biofilms on sHA disks were immersed in 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer and for 1 hour, postfixed with 1% OsO4 for 1 hour and dehydrated graded through ethanol series. The sample were dried by critical point drying and coated with gold using a sputter coater and viewed using S-4700 field emission scanning electron microscope (Hitachi High Technologies America, Inc.) [14].
As being visualized by scanning electron microscope (SEM) after mouth rinses were treated for 5 min on preformed biofilms sHA disks, the effects of EO ingredient were confirmed (Fig. 3B). The biofilms were treated with saline (Fig. 3A).
7. RNA isolation
P. intermedia ATCC 49046 old biofilms for 48 hours precultured into polystyrene 12-well was added to 500 μl of EO and saline respectively. The plates were incubated for 5 min at room temperature. Cell pellet was frozen by liquid nitrogen and crushed using a mortar and pestle. Total RNA was isolated using Trizol reagent (Invitrogen, Burlington, ON, Canada). RNA quality was assessed by Agilent 2100 bioanalyzer using the RNA 6000 Nano Chip (Agilent Technologies, Amstelveen, The Netherlands), and RNA quantification was performed using ND-2000 Spectrophotometer (Thermo Inc., DE, USA).
8. Library preparation and sequencing
For control and test RNAs, rRNA was removed using Ribo-Zero Magnetic kit (Epicentre, Inc., USA) from each 5 ㎍ of total RNA. The construction of library was performed using SMARTer Stranded RNA-Seq Kit (Clontech lab Inc., CA, USA) according to the manufacturer’s instructions. The rRNA depleted RNAs were used for the cDNA synthesis and shearing, following manufacture’s instruction. Indexing was performed using the Illumina indexes 1-12. The enrichment step was carried out using of PCR. Subsequently, libraries were checked using the Agilent 2100 bioanalyzer (DNA high sensitivity kit) to evaluate the mean fragment size. Quantification was performed using the library quantification kit using a StepOne Real-Time PCR system (Life technologies, Inc., USA). Highthroughput sequencing was performed as paired-end 100 sequencing using HiSeq 2500 (Illumina, Inc., USA).
9. Data analysis
Bacterial-Seq reads were mapped using Bowtie2 software tool in order to obtain the alignment file. Differentially expressed gene were determined based on counts from unique and multiple alignments using EdgeR within R (R development Core Team, 2016) using Bioconductor [15]. The alignment file also was used for assembling transcripts. Quantile normalization method was used for comparison between samples. Gene classification was based on searches done by DAVID. Statistical analysis was performed with SPSS Statistics program (ver. 24.0; IBM Corp., Chicago, IL, USA). Data were analyzed using a one-way ANOVA followed by Dunnett’s test.
Results
1. Ingredient and pH of mouth rinse solution
The pH and ingredients of mouth rinse solution were shown in (Table 1). The ingredients were menthol, eucalyptol, methyl salicylate, thymol, Benzethonium chloride (BC), isopropyl methlphenol, sodium fluoride, dipotassium glycyrrhizinate (DPZ), and chlorhexidine gluconate (CHX).
2. Mouth rinses treatment effects for preformed multiple oral biofilms
After mouth rinses, which are able to be purchased on the domestic market, were treated for 1 min on preformed multiple oral biofilms by 3 types of strain, ATP measured. ATP is an activated energy carrier that is present in all viable cells. When treated with mouthwash, there were significant differences compared with controls (P<0.01). It was treated by EO (No. 1), the viability of preformed oral biofilms were by 37.9% as the lowest. CHX (No. 4) was by 39.3%, as shown in (Fig. 1). The BC (No. 2) and DPZ (No. 3) were by 57.6%, 59.5%, respectively.
As shown in (Fig. 2), after mouth rinses were treated for 1 min on preformed multiple oral biofilms was serially diluted in TSA broth and plated on TSA and CFU were counted. The number of bacteria was reduced when exposed EO (No. 1) (2.7×109) than saline (No. 5) (2×1010). In CFU measured result comparing the controls, preformed multiple oral biofilms were reduced by the lowest value 39.0% up to 95.7% (P<0.05); especially, EO and CHX ingredient showed high antibacterial effects as 86.7% and 95.7%, respectively (P<0.001). EO is as high as CHX high antibacterial effects were shown on oral biofilms. The number of bacteria was reduced BC (No. 2) and DPZ (No. 3) by 54.6% (9.3×109), 39.0% (12.5×1010), respectively than control. Antibacterial effects were indicated in all products, but there were considerable differences according to products. The results are expressed as the Mean±SD.
Fig. 1
Viability of remaining multiple oral biofilms was determined by ATP and expressed in relative light units (Luminence). ATP-based luminescence quantification was performed in at least two independent experiments and expressed as the mean (SD) percent relative light units (RLU) versus saline-treated biofilms (Control). Different letters indicate significant differences among the biofilm conditions (P<0.05). *Image is numbering 1: EO, 2: BC, 3: DPZ, 4: CHX, 5: Saline
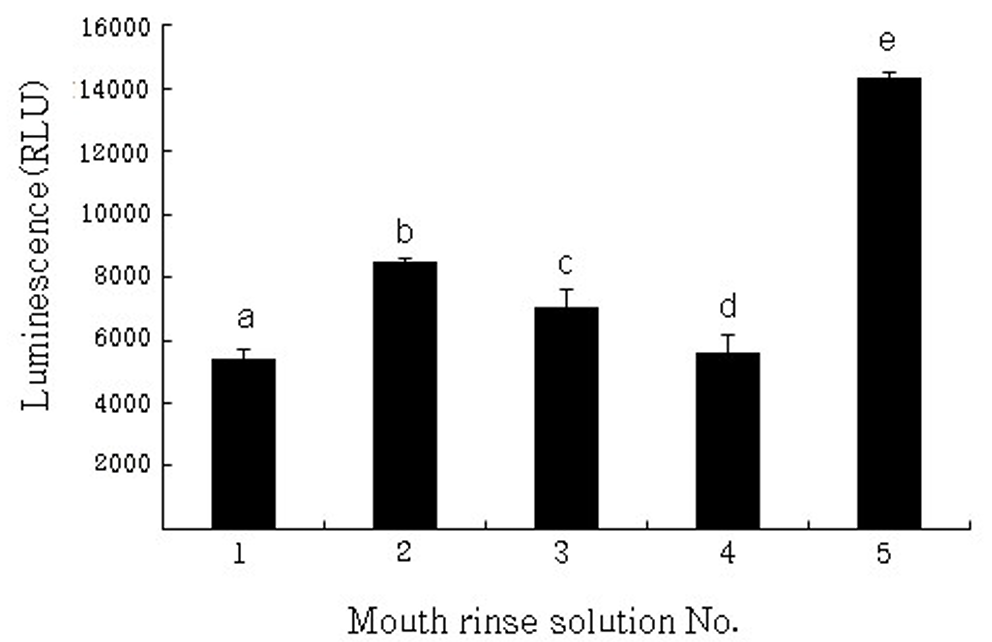
Fig. 2
Effect of mouth rinsing solution on the viability in multiple oral biofilms CFU was performed in at least three independent experiments and expressed as the mean (SD) percent colony form uint (CFU) versus saline-treated biofilms (Control). Different letters indicate significant differences among the biofilm conditions (pP<0.05). *Image is numbering 1: EO, 2: BC, 3: DPZ, 4: CHX, 5: Saline

3. Morphology visualized by SEM after EO
Scanning electron microscope (SEM) was used to observe the bacteria in the biofilms. As there are many studies on chlorhexidine, including experimental work on patients, effects using single cell biofilms and genetic screening, this study focused on EOs that are as effective as chlorhexidine and carried out SEM. As visualised by SEM after mouthwash treatment for 5 min on preformed biofilms sHA discs, the effects of the EO component or saline were confirmed (Fig. 3A, 3B). We observed a multilayered structure of well-formed bacterial biofilms were seen in the control group (Fig. 3A). The bacterial cells after EO were change size and some ruptured and small pieces of cell debris (Fig. 3B).
4. Results of RNA-sequence
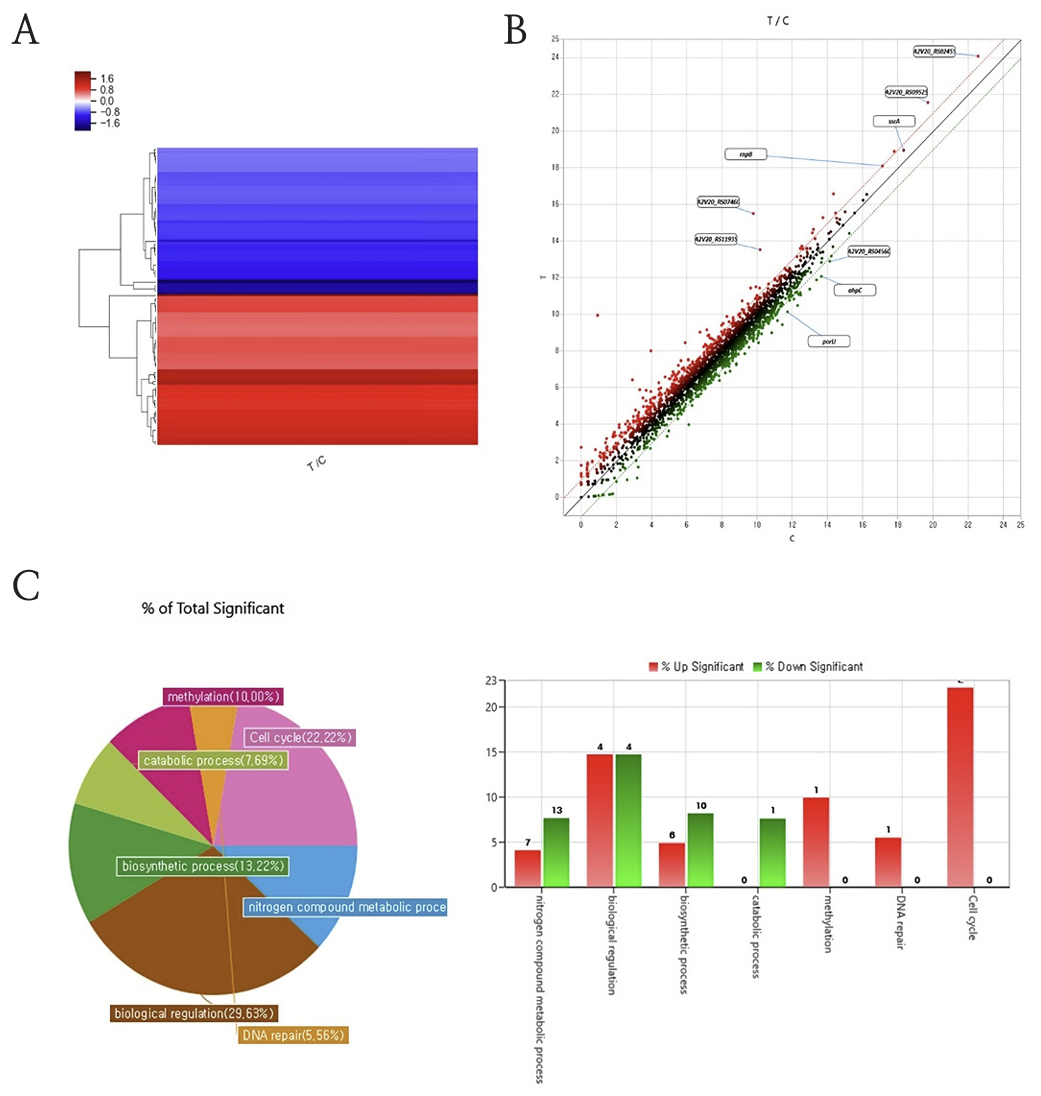
The effect of mouth rinse on oral biofilms was confirmed by global RNA sequencing. The biofilms cultured on 48 hour was with mouth rinse for 1 min then isolated RNA for prepare for the RNA seqencing. From the sequencing results, the different gene cluster was observed between mouth rinse treat group (T) and control (C). Also, gene expression in P. intermedia ATCC 49046 significantly altered in RNA transcription and protein translation related genes following essential oil (Fig. 4A, 4B). Moreover, the gene category chart displayed the number of genes with a significant difference in expression among each gene ontology. The scatter plot shows the gene changes between mouth rinse co-culture group and the control. Each dot represents an individual gene. Genes showing significant differential up-regulated expression in the RNA-seq data are highlighted in red, genes that are down-regulated highlighted in green. Moreover, the variation genes were categorized based on their culture condition (Fig. 4C).
Fig. 4
RNA sequencing results. The mouth rinse co-culture group (T) showed a significant difference between control group (C). (A) Clustering heatmap comparing the fold changes of genes between experimental groups; (B) A scatterplot to assess manifestation of the experimental control groups; (C) A chart showing the percentage and number of genes; the expression of each gene significant changed (below two images). *Abbreviations: (T) Treat group, (C) Control group
Discussion
Periodontitis and caries are infectious diseases of the oral cavity in which oral biofilms play a causative role [15]. Bacteria such as Streptococcus, Actinomyces, Fusobacterium and Prevotella were found in gingival crevice plaques. Fusobacterium is the bridge between the early stage and the late stage of oral biofilms [16]. As revealing the efficacy of mouth rinses other than tooth brushing for oral germs removal, further studies about mouth rinses have been actively progressed [17]. As ingredients in mouth rinses, there are EO (thymol, eucalyptol, menthol), bis-biguanides (CHX), quaternary ammonium compounds (CPC), domiphen bromide (BC), herbal extracts (sanguinarine), halogens (fluoride; iodine), and oxygenating agents (peroxides) etc [18]. Some reported that mouth washing with essiontial oil for 30 seconds resulted in decrease of viable bacterial counts in the saliva [19,20].
Most of the studies were conducted only with long terms and planktonic cells. There was a lack of research to shows that bacteria were reduced multiple oral biofilms as directed by the manufacturer. In this study, we examined the effect of mouth rinse solution against multiple oral biofilms cells, a major oral pathogen. All the commercial mouth rinse solutions were used for commercial use, and we shows that their effects after the formation of biofilms. For the 48h biofilms formation the mouth rinse solution significantly reduced the number of CFU/ml. Previous studies reported mouth rinses containing DPZ was used for 4 weeks, supragingival plaque and gingivitis were lessened [21]. Mouth rinses containing BC were effective on antiplaque and effective in killing S. mutans biofilms [7,22]. The oral solution containing BC or DPZ used in this study was reduced preformed multiple oral biofilms (Fig. 1, 2). However, CHX and EO had more effect on preformed multiple oral biofilms. CHX has more efficacy to restrain plaque if it has been used for 6 months, plaque could be decreased by 45% and gingivitis could be decreased by 80% to the maximum [23]. CHX has inhibitory effect gingivitis and dental plaque and growth S. mutans and Lactobacillus and ability to maintain low plaque scores and gingival health during for 3 weeks period [24]. Some studies shown that CHX can inhibit mitochondrial activity and protein and DNA synthesis and cause mitochondrial disorder, an increase in intracellular Ca2+, or cell death or necrosis by oxidation [25,26]. According to some research papers, within 3 weeks thickness of biofilms is more killing to CHX than biofilms and nutrient-limited biofilms [24] and statistically signigicant reduced in microorganisms when EO and CHX solutions were used for biofilms control [27,28]. Previously study reported CHX exerts cytotoxic effects on human periodontal tissues and interfering with periodontal regeneration [27].
EO was effect more predominantly through an anti-inflammatory process [29]. EO formulation was long-term control of gingival inflammation similar CHX and it was alternative to CHX respect to gingival inflammation [28]. In this study were obtained Similar results that EO was reduced ATP (37.9%) and CFU (86.8%) on the preformed multiple oral biofilms similar CHX. EO is as high as CHX high antibacterial effects were shown on preformed multiple oral biofilms than other oral rinses. The EO and CHX solution had similar antibiofilms effects. If EO has been used for a long term, plaque could be decreased by 56% and gingivitis was decreased by 35% [23]. EO commonly used formulations consist of thymol, eucalyptol, menthol, and methyl salicylate, demonstrated their efficacy against plaque and gingival inflammation [30]. Essential oil contain alcohol, so further research on alcohol free EO is needed.
In this study, we performed SEM and RNA analysis, focusing on essential oil that were as effective as chlorhexidine. The SEM of the preformed multiple oral biofilms confirmed surface structural change. As being morphology visualized by SEM after EO were treated on preformed biofilms sHA disks, the effects of EO ingredient were confirmed. This images show changes on the bacterial structure when treated with EO. SEM observation revealed that multiple oral biofilms surface that were treated with saline, showing a multilayered structure (Fig. 3A). The multiple bacterial cells with EO became unequal in size and some ruptured and divided (Fig. 3B).
RNA sequencing allows quantitative measurement of expression levels of genes and their transcripts. Moreover, the oral biofilms gene expression changes due to mouth rinses were confirmed with RNA sequencing. The genes which expression between control (C) and mouth rinse treat (T) groups made a gene cluster. From these results, the mouth rinses can change the gene expression not just killed the bacteria. Also, specific gene expressions were displayed in gene expression scattering graph. The histidine kinase related genes (ssrA) and RNase (rnpB), ribosome associated inhibitor (raiA), ribosome recycling factors (rrf) were over expressed by mouth rinse. The gene clustering indicated a different gene expression between control and mouth rinse treat groups (Fig. 4A). The results indicate that the down regulation of genes, including translating phase genes (porU, ahpC). The ahpC gene protects cells against oxidative stress by detoxifying peroxides (Fig. 4B). However cell cycle and methylation genes were up regulated. These gene variations suggest that DNA methylation is a crucial part of general organismal evolution and cellular variation (Fig. 4C). From these results, the mouth rinse regulates the bacteria’s biomolecules and finally showed anti-bacteria effect. Additionally, the changed gene expressions were categorized and the biological regulation genes, cell cycle related genes and nitrogen compound metabolic process genes were changed due to mouth rinse.
In this study, the effects of mouth rinse solutions studied in planktonic cells and single cell biofilms were confirmed in young oral biofilms, and the morphological and RNA changes in biofilms after treatment with mouth rinse solutions are of great significance. However, the limitation of this study is that the effect of the mouth rinse solution on young biofilms mixed with only three strains cannot be considered as the effect of all strains in the oral cavity where many strains live.
In addition, the SEM and RNA results are limited to the results of mouthwashes with essential oil with good efficacy, and further studies of mouthwashes on mature biofilms are needed. The further study is needed to investigate the mechanism of effect of mouth rinse.
Conclusions
In this study, we examined the antimicrobial effects of oral mouth rinses with different compositions on long-established oral biofilms containing various oral microorganisms. Based on our findings, we obtained the following conclusions.
1. EO reduced the viability of oral biofilms by 37.9% as the lowest (P<0.01).
2. EO is as high antibacterial effects were shown on oral biofilms in the counted colony forming units (CFU)(P<0.001).
3. SEM was used to observe the bacterial cells after EO were change size and some ruptured and small pieces of cell debris.
4. The gene expression of these two groups was significantly change in RNA transcription, and protein translation related genes.
In conclusion, mouthwash with different ingredients had different effects, with CHX and EO being the most effective. SEM of the preformed multiple oral biofilms confirmed the change in surface structure. Mouth rinse can alter gene expression and showed specific gene expression of the bacteria matched RNA sequencing.